DNA Traffic Jams: Meet KCTD10, the Guardian of Genome Safety!
In every cell of your body, a sophisticated molecular traffic flow takes place: the replication machinery (replisome), like a high-speed sports car, rapidly copies DNA to ensure genetic information is not lost during cell division; the transcription machinery (RNA polymerase), like a heavy truck, slowly reads genes and synthesizes RNA. However, these two majors "production lines" operate on the same DNA "lane." A collision can trigger transcription-replication conflicts (TRCs)-a major cause of DNA damage, gene mutations, and even cancer. Approximately 70% of human tumors are associated with genomic instability, and TRCs are one of the key drivers behind this.
Now, a Mayo Clinic team, publishing paper in Nature, has identified a solution to DNA "traffic jams"-the protein KCTD10. Like a precise traffic commander, it can promptly clear conflicts, safeguard genomic stability, and potentially open up new avenues for cancer treatment.
The "two-way crisis" of DNA replication and transcription is more complex than imagined. The replication machinery can move thousands of bases per second, while the transcription machinery moves at only one-tenth that speed. If the two travel in the same direction on the same DNA strand, the replication machinery can easily "catch up" with the transcription machinery. Even more dangerous, in the human genome, DNA replication origins are often concentrated in regions of active transcription. This means that "traffic rush hour" and "narrow lanes" are combined, greatly increasing the probability of TRCs. Scientists have known that TRCs can cause DNA breaks and mutations, but how cells "sense" this conflict and "manage" it in a timely manner to avert disaster has remained a mystery.
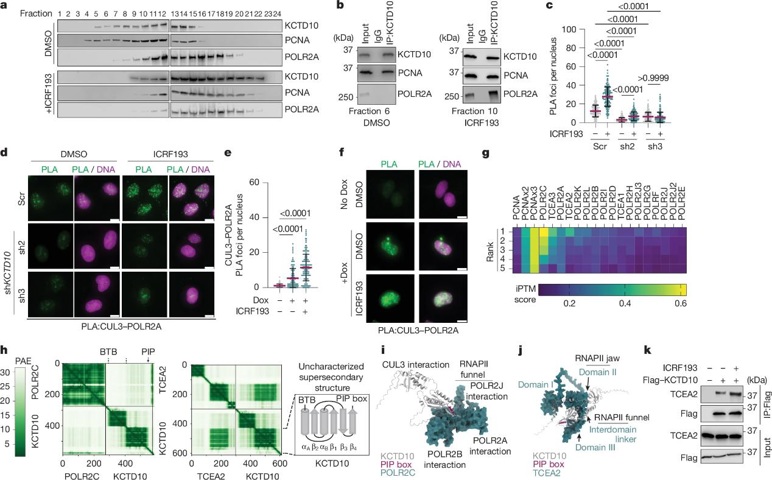
The discovery of KCTD10 fills this gap. The team discovered that KCTD10 possesses a unique ability-it can simultaneously "hold hands" with both the replication and transcription machinery: it binds to the key replication protein PCNA through its C-terminal PIP box and interacts with RNA polymerase II (POLR2A) in the transcription machinery, acting as a "molecular bridge" bridging the two systems. When the replication machinery is about to catch up with the transcription machinery ahead, KCTD10 quickly becomes alerted and recruits the CUL3 protein through its BTB domain, forming the CUL3-KCTD10 E3 ubiquitin ligase complex. This complex acts like a traffic enforcement team, attaching a ubiquitin tag to TCEA2, a key factor in the transcription machinery-essentially signaling TCEA2 to pull over. TCEA2 is then cleared by the cell's degradation system, and the transcription machinery disassembles, clearing the way for the replication machinery. The entire process is efficient and precise, preventing DNA collisions.
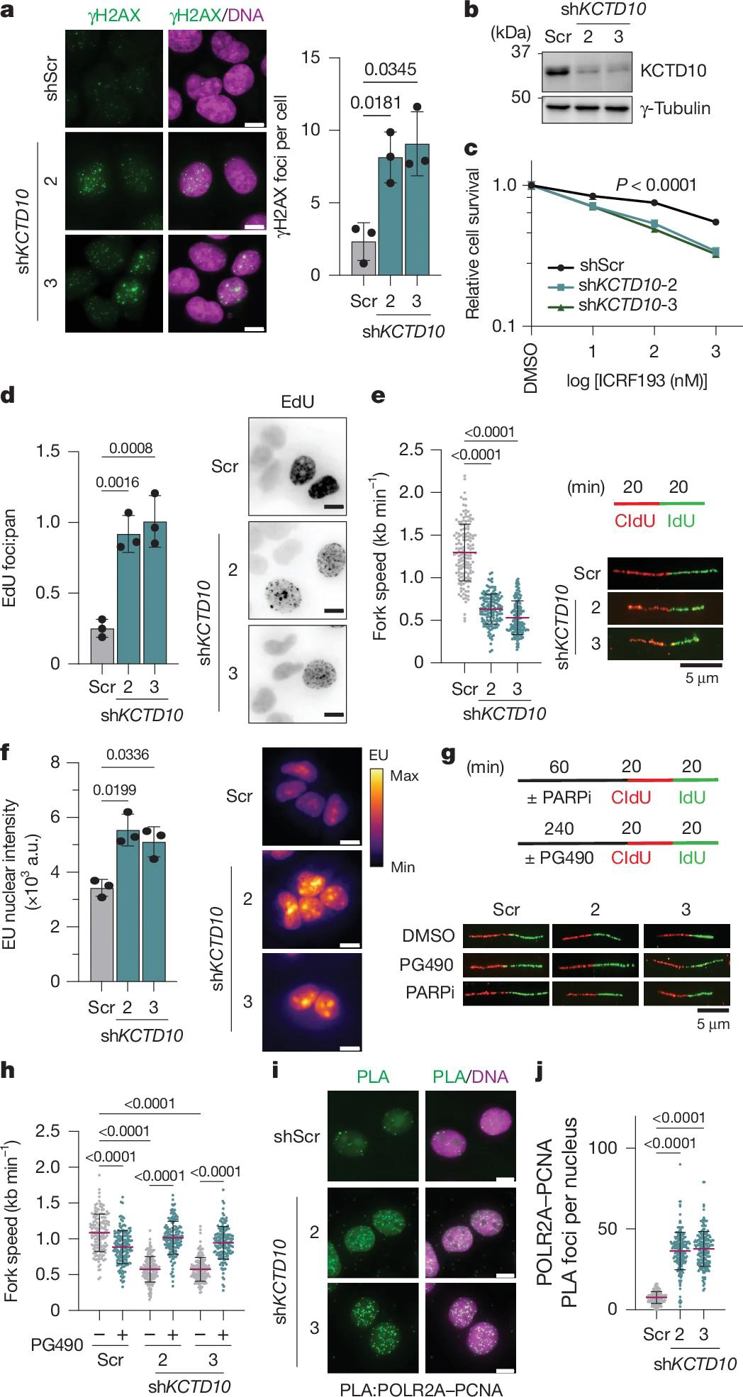
Even more ingeniously, KCTD10 only targets "co-directional conflicts". There are two types of TRCs on DNA: co-directional (replication and transcription in the same direction) and anti-directional (opposite directions). Previously, anti-directional conflicts were believed to be more harmful, but research has found that co-directional conflicts, if not resolved promptly, can also cause severe DNA damage. KCTD10, like a "recognition radar", precisely locates co-directional conflict sites by analyzing replication fork orientation (RFD) and transcription direction. CUT&RUN sequencing revealed that 77% of KCTD10-bound sites are co-directional TRCs. Furthermore, the number of co-directional conflict sites increased sevenfold when induced by DNA topoisomerase inhibitors such as ICRF193, further demonstrating that it specifically targets these "traffic jams".
Without KCTD10, cells become paralyzed. When researchers knocked out KCTD10 in cells, TCEA2 could not be promptly cleared, blocking the transcription machinery on DNA. This led to a three-fold increase in the incidence of TRCs, a significant increase in γH2AX (a marker of DNA damage), and a doubling of the number of micronuclei (a hallmark of genomic instability) within the cells. Over time, this genomic instability can lead to the accumulation of mutations and accelerated tumor formation. Even more surprising, KCTD10 deficiency also causes developmental delay in mice, demonstrating that it is not only a cancer prevention factor but also a necessary factor for normal physiological activities.
The clinical value of this study opens a new window for anti-cancer treatment. The loss of KCTD10 expression in many cancer cells renders their genomes highly vulnerable. They are unable to effectively activate TRCs, and their rapid division requires extensive DNA replication, akin to adding traffic to a chaotic traffic flow, making them more susceptible to DNA damage. Researchers have found that KCTD10-deficient cancer cells are more sensitive to topoisomerase II inhibitors (such as ICRF193) and ATM/CHK2 inhibitors. The former exacerbates TRCs, while the latter blocks DNA damage repair. This double-edged attack increases cancer cell death. This suggests that KCTD10 could serve as a new biomarker, helping physicians identify tumors suitable for these targeted therapies and precisely narrow the therapeutic window.
The research team has incorporated this discovery into the Mayo Clinic's "Precure Project", which will subsequently identify cancer types (such as some lung and colon cancers) with high KCTD10 deletion rates and develop targeted drugs. For example, drugs could be designed to enhance KCTD10 activity, helping normal cells resist TRCs. Alternatively, a topoisomerase inhibitor and an ATM inhibitor could be combined to target KCTD10-deficient cancer cells, amplifying their genomic vulnerabilities and achieving precise targeting.
Reference
- Kloeber, Jake A., et al. "KCTD10 is a sensor for co-directional transcription-replication conflicts." Nature (2025): 1-10.

Your email address will not be published. Required fields are marked *